With the help of plastimatch you can convert any rt-structure file into binary image by running:
plastimatch convert --input RS.dcm --output-prefix masks --prefix-format nrrd --output-ss-list image.txt
Now, if you wish to have the exported binary images of each structure registered with your rt-dose file, you can run:
plastimatch convert --input RS.dcm --output-prefix masks --prefix-format nrrd --output-ss-list image.txt --input-dose-img "RD.dcm"
The plastimatch will generate the binary images in .nrrd format (also .mha can be specified in –prefix-format mha – mha is the default option, by the way -). The generated files can be found under the directory called “masks“.
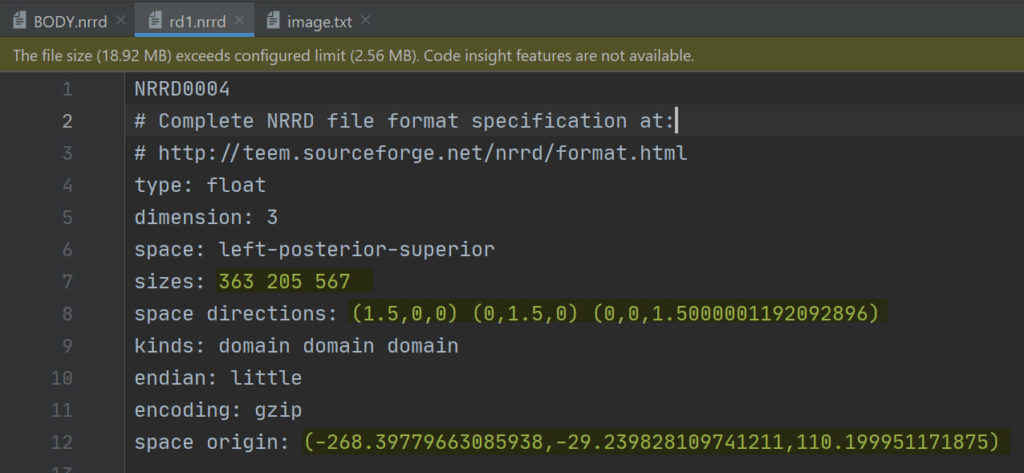
Looking also in the file header of a binary image and the RT-Dose file, you can see that the space origin, size and space directions in the two files are the same.
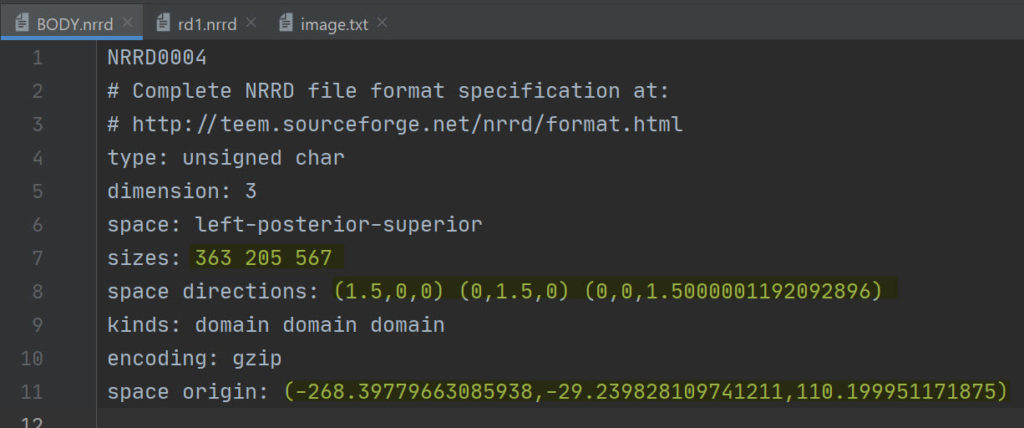
To read the nrrd file, you can use SimpleITK python package, as shown in the code snippet below:
import SimpleITK as sitk
import pylab as plt
import nrrd as nr
import numpy as np
def main():
file_body = "./Prostata_1-IntraObserver/Sum-2/masks/BODY.nrrd"
img_body = sitk.ReadImage(file_body)
mask_body = sitk.GetArrayFromImage(img_body)
nrrd_mask_body = nr.read(file_body)
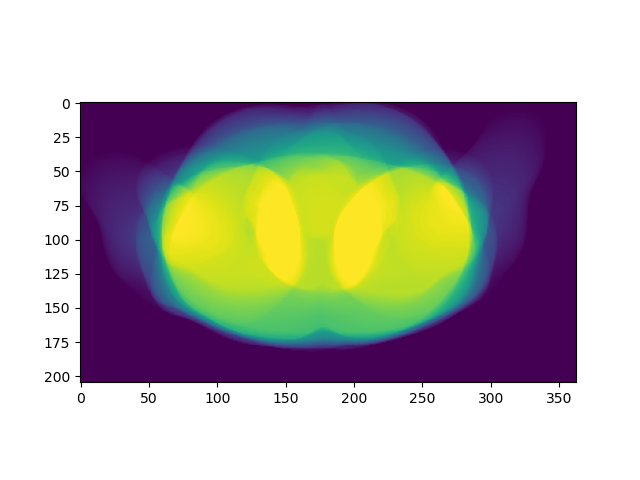
plt.imshow(np.sum(mask_body, axis=0))
plt.show()
dosefile = "./Prostata_1-IntraObserver/Sum-2/RD_PC1+PC3-2.dcm"
dose_img = sitk.ReadImage(dosefile)
arr_dose = sitk.GetArrayFromImage(dose_img)
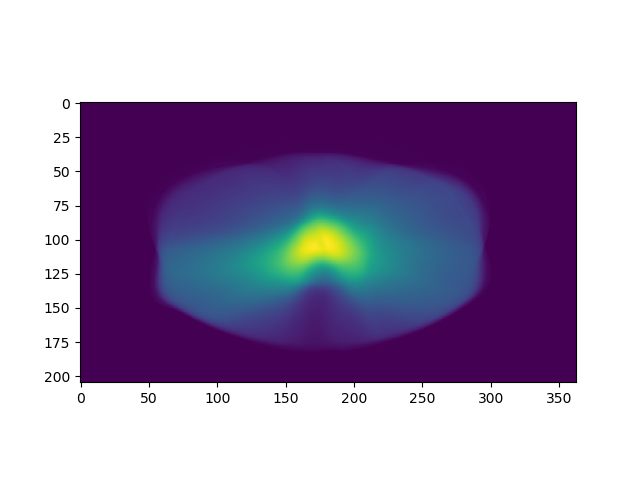
plt.imshow(np.sum(arr_dose, axis=0))
plt.show()
if __name__ == '__main__':
main()
The results are two figures: